-
How to use the 'Druglikeness Evaluation'?
-
How to use the 'ADMET Prediction'?
-
How to use the 'Systematic Evaluation'?
-
How to use the 'Database'?
| How to use the 'Druglikeness Evaluation'?
This Druglikeness analysis module is designed for users to evaluate the druglikeness profiles of a batch of chemical entities and it includes 5 commonly used druglikeness rules and 1 specific predictive model built by us.
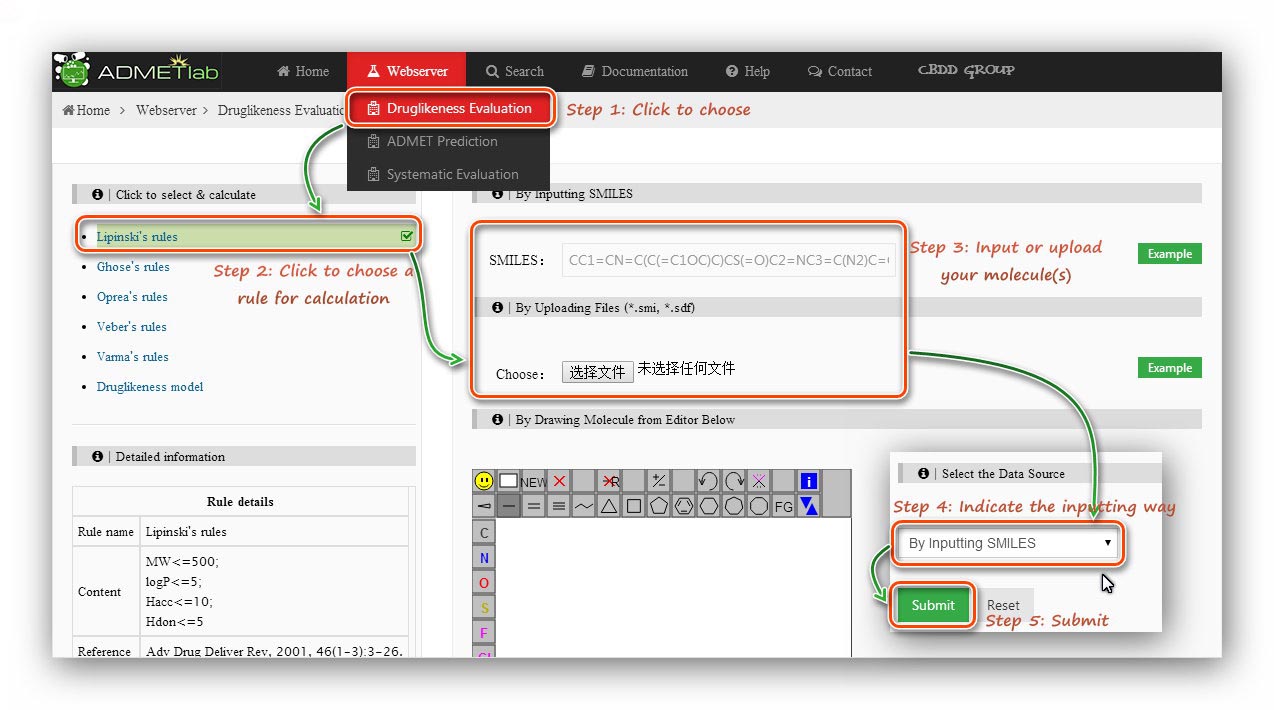
Step 1: Click to go to index page.
Step 2: Click to choose a rule/model to calculate.
Step 3: Users are required to submit a molecular structure, SMILES or a molecular file (*.sdf).
Here we use a *.sdf file to calculate.
Step 4: Indicate the inputting way.
Step 5: Processing and results.
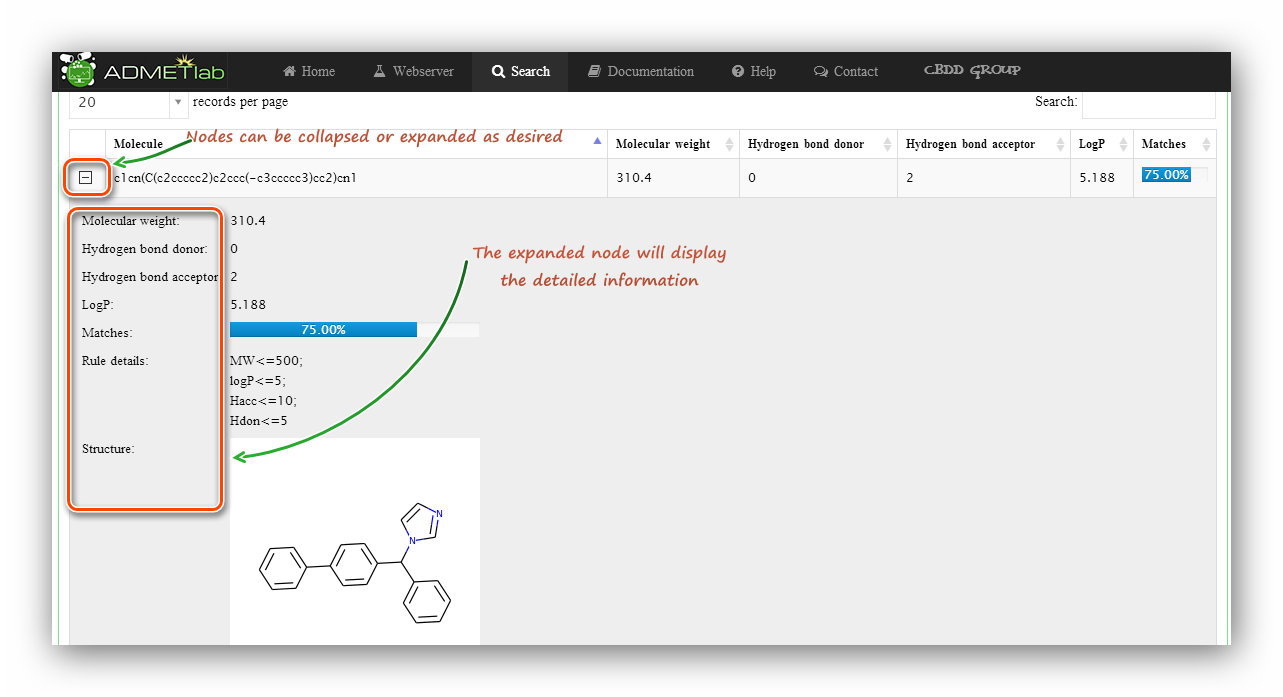
Click to submit. The processing will last for a while and then users will be redirected to the result page. The result page firstly gives a statistic about molecule numbers, Then provides a ‘Download’ button to download a *.csv file containing the values. A datatable will show detailed information of the calculating results. A matching percentage of druglikeness rules or a predictive category with its probability value and corresponding molecular physicochemical properties will be displayed in the table. Besides, the ‘Searching’ and ‘Ranking’ functionalities are also available for users to look for a certain item in the results and get an increasing or decreasing order of the values.
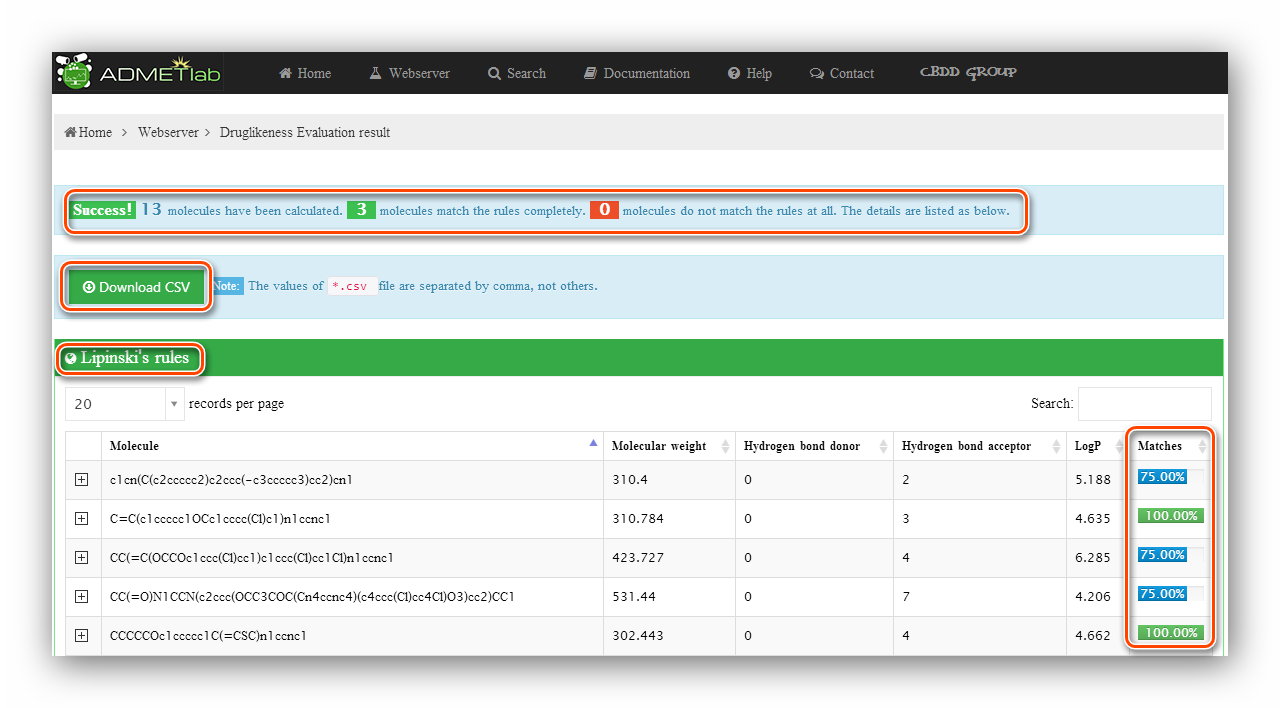
What’s more, every item gets a node, click to expand the node, users will get more detailed information about the item like molecular structure.
| How to use the 'ADMET Prediction'?
The ADMET evaluation module can be used to further assess specific pharmacokinetic properties by using 9 regression models and 20 classification models with good performance available in this web server.
Step 1: Click to go to index page.
Step 2: Click to choose a property to predict. When clicked, the sidebar on the right will display the detailed information about the model.
Step 3: Users are required to submit a molecular structure, SMILES or a molecular file (*.sdf).
Here we use a *.sdf file to calculate.
Step 4: Indicate the inputting way.

Step 5: Processing and results.
Click to submit. The processing will last for a while and then users will be redirected to the result page.
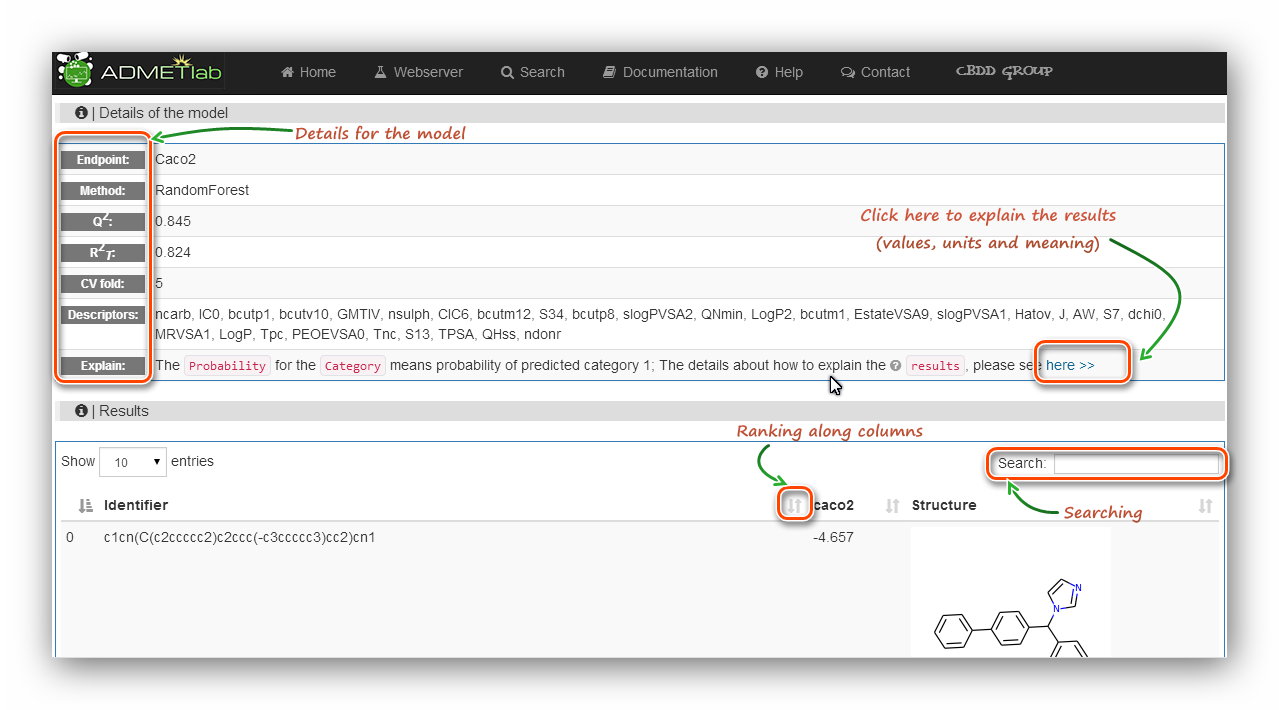
The ‘Details of the model’ section reminds users what they are calculating.
The ‘Results’ section provides ‘Identifier’ (Molecular SMILES),
‘Endpoint’(Property), ‘Structure’ of each item. Also the ‘Ranking’
and ‘Searching’ tools are also available for users. It should be noted that in the result table,
there is no unit or reference about the endpoint, and users should click the ‘here’ link
behind the ‘Explain’ cell to get the related explanation.
| How to use the 'Systematic Evaluation'?
This module allows users to evaluate every stage of ADME/T process of one molecule.
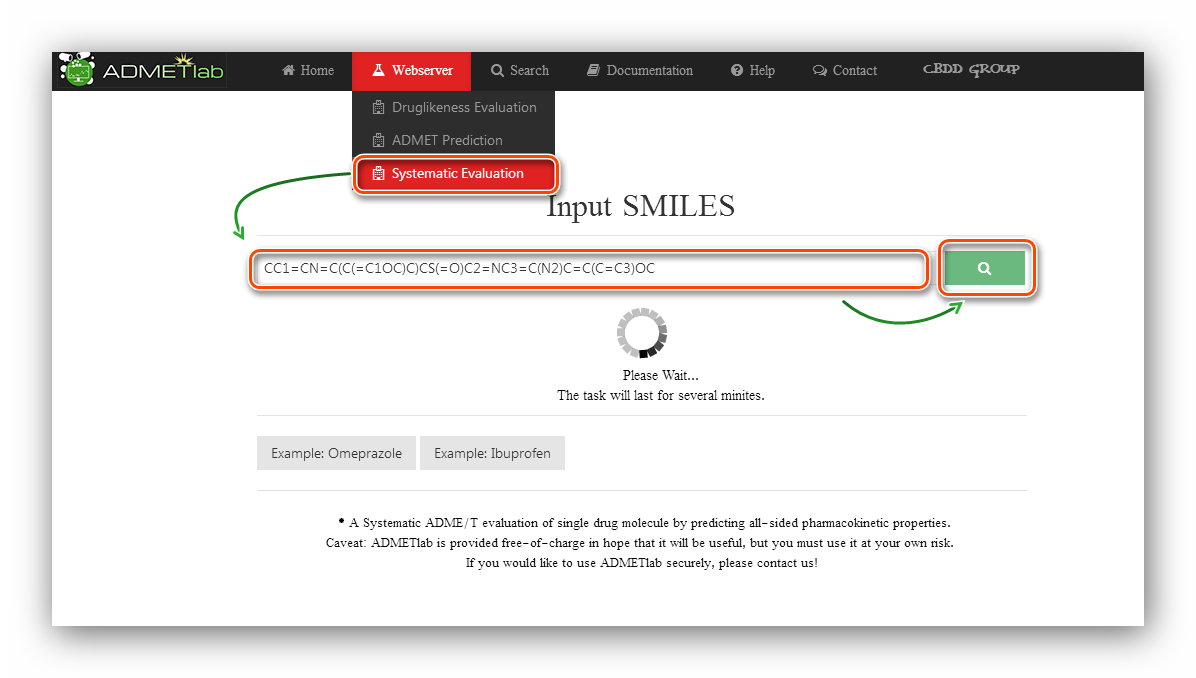
Step 1: Click to go to index page.
Step 2: Users are required to input SMILES.
Here we use the first example(Omeprazole) to calculate.
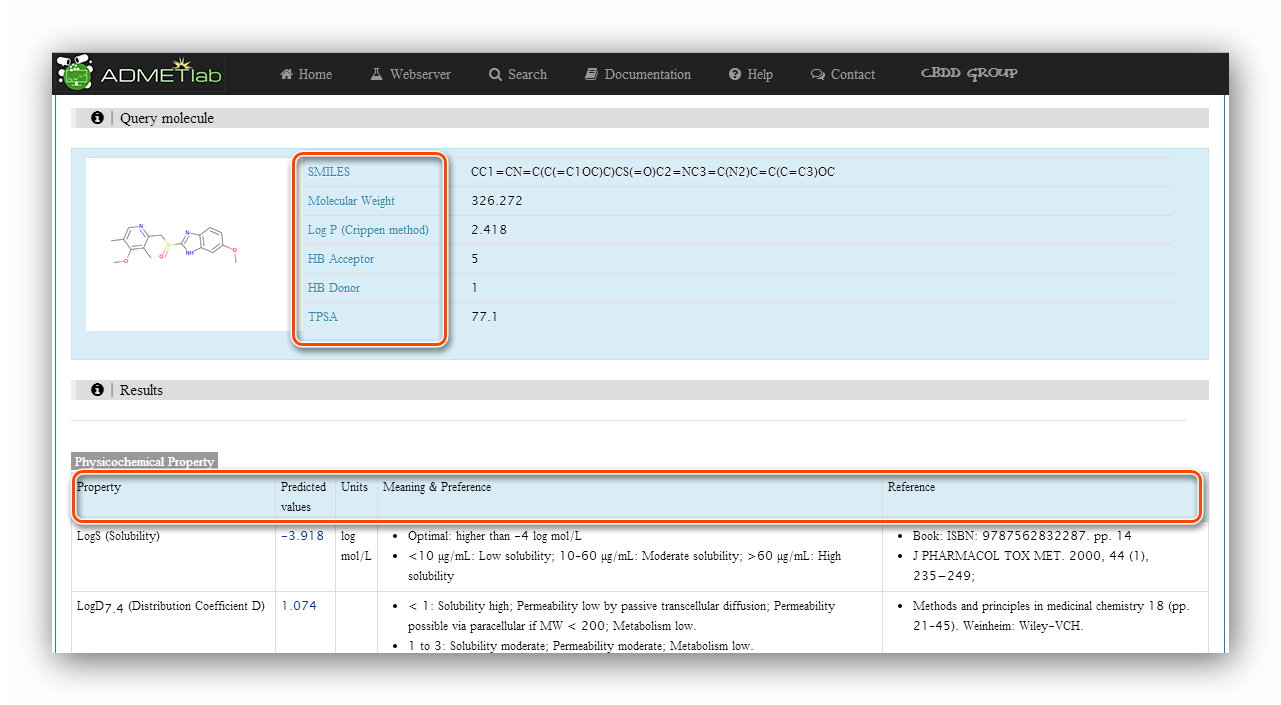
Step 5: Processing and results.
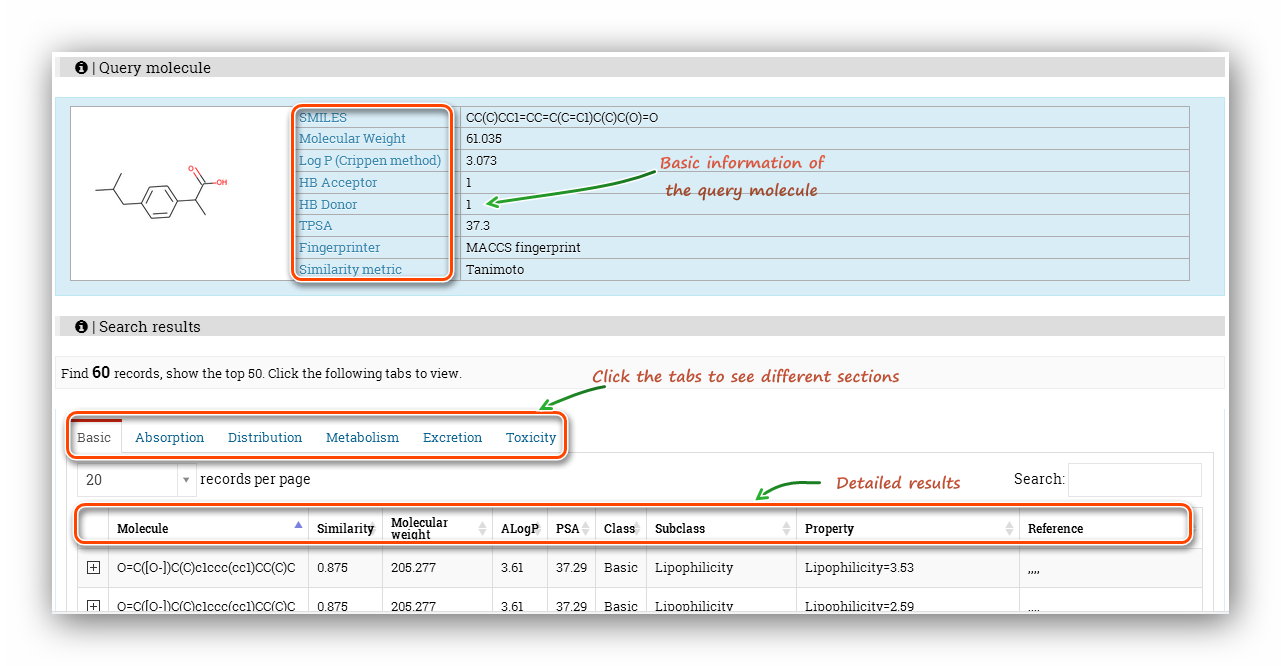
The 'Query molecule' section reminds users what they are calculating and provides some basic properties of the molecule. The results section displays 'Property'(Endpoint name), 'Predicted values', 'Units', 'Meaning&Preference' and 'Reference' of each endpoint.
| How to use the 'Database'?
This module provides three ways to search the database and further infer properties of the query molecule. They are 'Accurate Search', 'Range' and 'Similarity'.
Step 1: Click to go to index page.
Step 2: Input SMILES.
Here we use the example(Ibuprofen) to calculate.
Step 3: Set parameters. Here, we use 'MACCS' fingerprint and 'Tanimoto' metric. The similarity was set as 0.7.
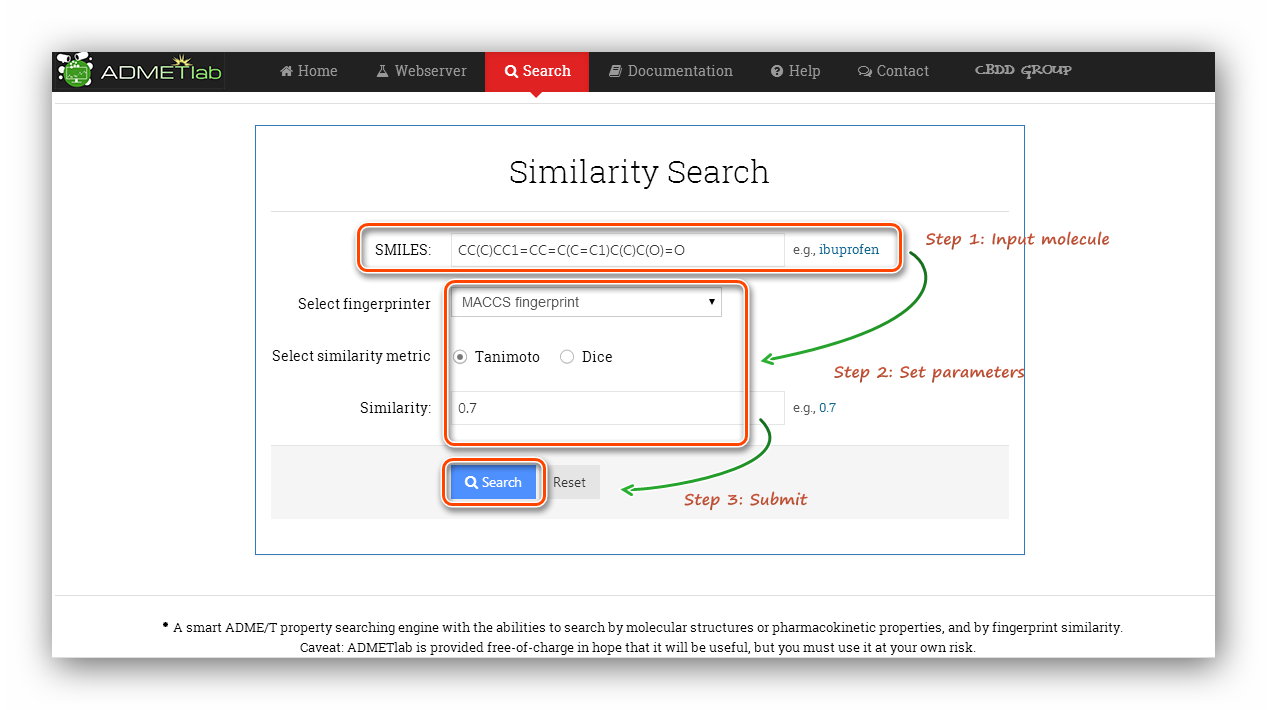
Step 5: Processing and results.
The 'Query molecule' section reminds users what they are calculating and provides some basic properties of the molecule. The 'Search results' section displays the 'Molecule'(SMILES), 'Similarity', 'Molecular weight', 'ALogP', 'PSA', 'Class', 'Subclass', 'Property', 'Reference'. Note that the ADMET was split into 6 tabs to display, and users should click these tabs to see different items. Some items seems to be the same, actually, they are not. Expand the node, more detailed information will be displayed, which will help distinguish these items.